SNURF differentially methylated region
SNURF differentially methylated region#
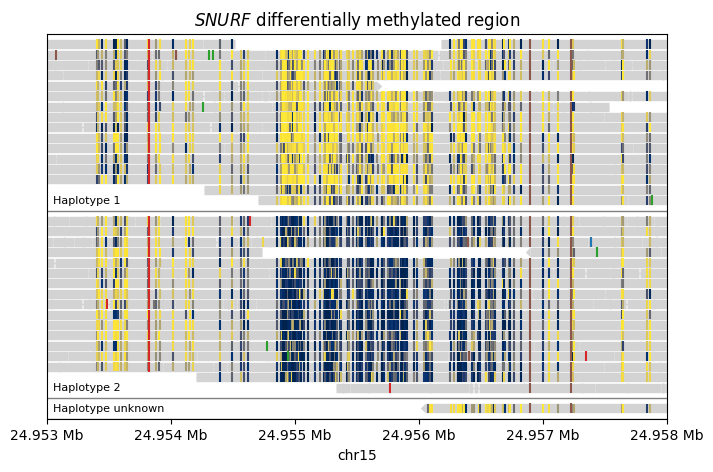
This example demonstrates the paternally imprinted differentially methylated region (DMR) in the SNURF gene. Usually this DMR is only methylated in the paternal haplotype but not in the maternal haplotype. We use PacBio HiFi long-read sequencing data generated from the HG002 individual from the Genome in a Bottle cohort. The BAM file used here has been haplotype-tagged using the HP tag.
Data sources:
PacBio HiFi sequencing: URL
import matplotlib.pyplot as plt
import lakeview as lv
CHROMOSOME = "chr15"
PACBIO_BAM_PATH = "../../tests/data/HG002_GRCh38_SNURF_haplotagged.bam"
painter = lv.SequenceAlignment.from_file(PACBIO_BAM_PATH, CHROMOSOME)
fig, ax = plt.subplots(figsize=(8, 5))
painter.draw_alignment(
ax,
group_by=lambda segment: segment.get_tag("HP") if segment.has_tag("HP") else 3,
group_labels={1: "Haplotype 1", 2: "Haplotype 2", 3: "Haplotype unknown"},
show_modified_bases=True,
modified_bases_kw=dict(
linewidth=1.5,
colormaps={("C", "m", "+"): "cividis", ("C", "m", "-"): "cividis"},
),
)
ax.set_title(r"$\it{SNURF}$ differentially methylated region")
ax.set_xlim(24.953e6, 24.958e6)
ax.set_xlabel("chr15")
ax.xaxis.set_major_formatter(lv.plot.BasePairFormatter("Mb", show_suffix=True))
Show code cell output
fig
