GAPDH Iso-Seq
GAPDH Iso-Seq#
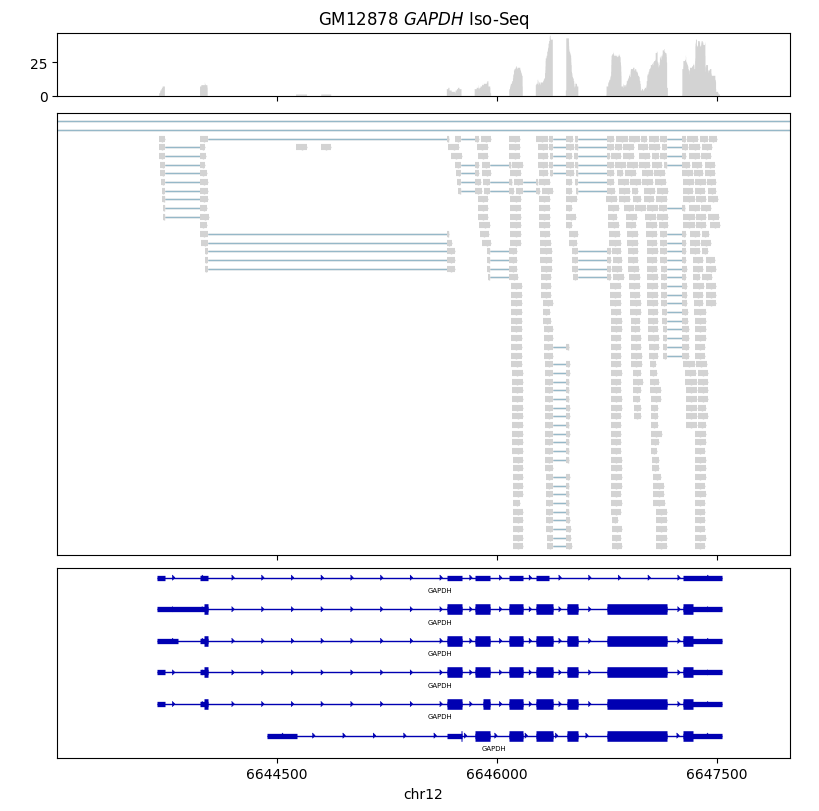
Here we visualize PacBio Iso-Seq RNA sequencing data for the GAPDH gene. The sequencing data was generated from the GM12878 lymphoblastoid cell line, which originated from the NA12878 individual from the 1000 Genomes cohort.
Data sources:
import gzip
import lakeview as lv
CHROMOSOME, START, END = "chr12", int(6.643e6), int(6.648e6)
RNA_BAM_PATH = "../../tests/data/GM12878_RNAseq_GAPDH.sample=0.002.bam"
REFSEQ_GFF_PATH = "../../tests/data/Refseq_GRCh37_genomic_annotation.gff.gz"
alignment_painter = lv.SequenceAlignment.from_file(RNA_BAM_PATH, CHROMOSOME)
with gzip.open(REFSEQ_GFF_PATH, "rt") as f:
annotation_painter = lv.GeneAnnotation.from_refseq(
f, region=("NC_000012.11", (START, END))
)
gv = lv.GenomeViewer(3, figsize=(8, 8), height_ratios=(1, 7, 3))
alignment_painter.draw_pileup(gv.axes[0], show_mismatches=False)
alignment_painter.draw_alignment(
gv.axes[1],
show_arrowheads=False,
show_soft_clippings=False,
show_hard_clippings=False,
show_mismatches=True,
max_rows=50,
show_group_labels=False,
show_group_separators=False,
)
annotation_painter.draw_transcripts(gv.axes[2], arrows_kw=dict(style="fishbone"))
gv.set_xlim(6.643e6, 6.648e6)
gv.set_xlabel(f"{CHROMOSOME}")
gv.set_title(r"GM12878 $\it{GAPDH}$ Iso-Seq")
Show code cell output
gv.figure
